Beginner `climr` workflow
climr_workflow_beg.RmdUsing the downscale() function
The basic use of climr is to pass a
data.table of point coordinates, IDs and elevation to
downscale() and select the type of climate projection or
values wanted - e.g. below we ask for future projections in two periods,
using two emissions scenarios and two General Circulation Models. We
also specify we want 3 individual runs of each model/scenario
combination, in addition to the ensemble mean (returned by default).
library(climr)
## provide a data.frame or data.table of point coordinates, IDs and elevation
my_points <- data.frame(
lon = c(-123.4404, -123.5064, -124.2317),
lat = c(48.52631, 48.46807, 49.21999),
elev = c(52, 103, 357),
id = seq_len(3)
)
## climr query for the data.frame
ds_out <- downscale(
xyz = my_points,
gcms = c("GFDL-ESM4", "EC-Earth3"), # specify two global climate models
ssps = c("ssp370", "ssp245"), # specify two greenhouse gas concentration scenarios
gcm_periods = c("2001_2020", "2041_2060"), # specify two 20-year periods
max_run = 3, # specify 3 individual runs for each model
ensemble_mean = FALSE, # don't return ensemble mean of individual runs
db_option = "local", # perform downscaling locally (instead of in database)
vars = c("PPT_an", "CMD_an", "CMI_an")
)
#> .
#> .
#> .The resulting output is a data.table, printed below:
| id | GCM | SSP | RUN | PERIOD | PPT_an | CMD_an | CMI_an |
|---|---|---|---|---|---|---|---|
| 1 | NA | NA | NA | 1961_1990 | 939.1875 | 318.3206 | 41.45086 |
| 1 | EC-Earth3 | ssp245 | r15i1p1f1 | 2001_2020 | 930.9058 | 347.4192 | 37.11035 |
| 1 | EC-Earth3 | ssp245 | r15i1p1f1 | 2041_2060 | 964.1574 | 375.0391 | 35.28234 |
| 1 | EC-Earth3 | ssp245 | r1i1p1f1 | 2001_2020 | 950.7793 | 316.7011 | 40.82642 |
| 1 | EC-Earth3 | ssp245 | r1i1p1f1 | 2041_2060 | 1052.9310 | 388.9063 | 42.02329 |
| 1 | EC-Earth3 | ssp245 | r4i1p1f1 | 2001_2020 | 921.4209 | 362.2182 | 34.79205 |
| 1 | EC-Earth3 | ssp245 | r4i1p1f1 | 2041_2060 | 944.4196 | 390.0571 | 31.17417 |
| 1 | EC-Earth3 | ssp370 | r15i1p1f1 | 2001_2020 | 931.8873 | 358.3112 | 35.62798 |
| 1 | EC-Earth3 | ssp370 | r15i1p1f1 | 2041_2060 | 1003.8444 | 358.1437 | 38.15592 |
| 1 | EC-Earth3 | ssp370 | r1i1p1f1 | 2001_2020 | 887.5156 | 338.4092 | 33.32164 |
| 1 | EC-Earth3 | ssp370 | r1i1p1f1 | 2041_2060 | 926.0189 | 376.1525 | 29.52610 |
| 1 | EC-Earth3 | ssp370 | r4i1p1f1 | 2001_2020 | 949.9003 | 342.5648 | 37.49923 |
| 1 | EC-Earth3 | ssp370 | r4i1p1f1 | 2041_2060 | 1043.8276 | 341.0114 | 43.32436 |
| 1 | GFDL-ESM4 | ssp245 | r1i1p1f1 | 2001_2020 | 939.8992 | 318.3508 | 40.16827 |
| 1 | GFDL-ESM4 | ssp245 | r1i1p1f1 | 2041_2060 | 977.5459 | 377.2941 | 35.35334 |
| 1 | GFDL-ESM4 | ssp245 | r2i1p1f1 | 2001_2020 | 923.5054 | 349.3078 | 36.74840 |
| 1 | GFDL-ESM4 | ssp245 | r2i1p1f1 | 2041_2060 | 1003.5399 | 383.7482 | 38.87800 |
| 1 | GFDL-ESM4 | ssp245 | r3i1p1f1 | 2001_2020 | 944.9775 | 345.9026 | 38.66984 |
| 1 | GFDL-ESM4 | ssp245 | r3i1p1f1 | 2041_2060 | 988.8887 | 383.6453 | 36.43479 |
| 1 | GFDL-ESM4 | ssp370 | r1i1p1f1 | 2001_2020 | 933.8684 | 331.5469 | 39.06149 |
| 1 | GFDL-ESM4 | ssp370 | r1i1p1f1 | 2041_2060 | 974.3047 | 361.3603 | 37.31249 |
Listing available options
The list_* functions (see ?list_gcm) are useful to see
available options, for example:
list_gcms()
#> [1] "ACCESS-ESM1-5" "BCC-CSM2-MR" "CanESM5" "CNRM-ESM2-1"
#> [5] "EC-Earth3" "GFDL-ESM4" "GISS-E2-1-G" "INM-CM5-0"
#> [9] "IPSL-CM6A-LR" "MIROC6" "MPI-ESM1-2-HR" "MRI-ESM2-0"
#> [13] "UKESM1-0-LL"
list_gcm_periods()
#> [1] "2001_2020" "2021_2040" "2041_2060" "2061_2080" "2081_2100"
list_ssps()
#> [1] "ssp126" "ssp245" "ssp370" "ssp585"These functions can be used as arguments in downscale().
For example, vars = list_variables() could be specified to
obtain data for all available variables, or
gcms = list_gcm()[c(1,4,5,6,7,10,11,12)] could be used to
specify the eight-gcm ensemble recommended by Mahony et al. (2022).
Working with raster data
A common use of climr is to obtain climate data for a
user-provided raster grid. Here is some sample code showing how to do
so. climr now supports native raster downscaling, so we can
simply pass in a 1-layer SpatRaster containing elevation
values, and climr will return a raster stack with the same
extent and resolution as the input, where each layer corresponds to a
variable.
library(terra)
library(data.table)
## get the sample digital elevation model (dem) provided with `climr`
dem_vancouver <- get(data("dem_vancouver")) |>
unwrap()
## A simple climr query. This will return the observed 1961-1990 and 2001-2020 mean annual temperature (MAT).
ds_out <- downscale(
xyz = dem_vancouver,
obs_periods = "2001_2020",
gcms = list_gcms()[4],
ssps = list_ssps()[2],
gcm_periods = "2041_2060",
vars = c("MAT")
)
#> .
#> .
plot(ds_out, range = c(2,14))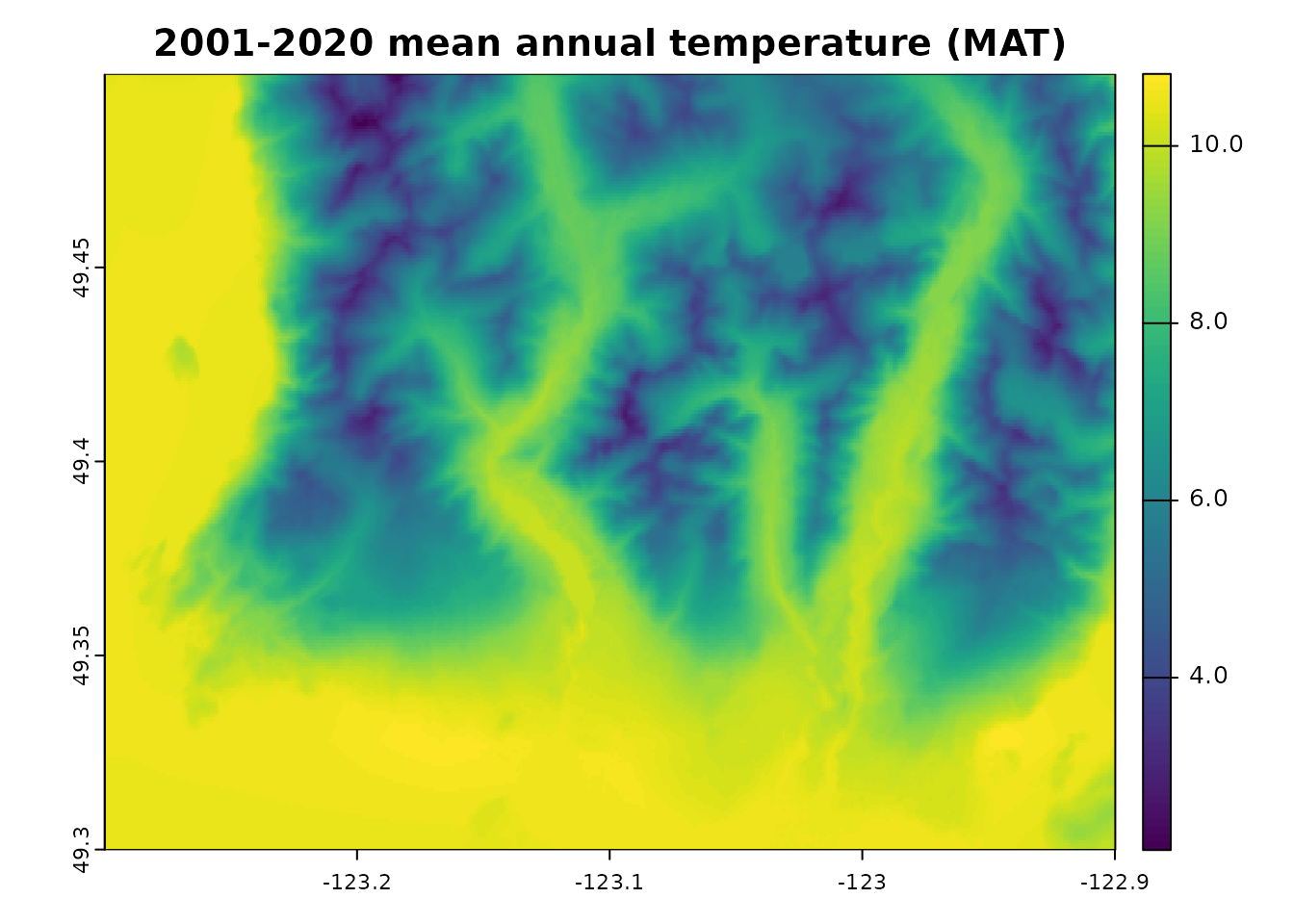
Looking under the hood
You can download and investigate the raw data that climr
uses for downscaling, such as the reference climatological maps, the
observational time series, and the GCM anomalies. See the section
“Workflow with *_input functions and
downscale” in
vignette("vignettes/climr_workflow_int.Rmd") for an
example.