Fits one or more distributions to species sensitivity data.
Usage
ssd_fit_dists(
data,
left = "Conc",
...,
right = left,
weight = NULL,
dists = ssd_dists_bcanz(),
nrow = 6L,
rescale = FALSE,
odds_max = 0.999,
reweight = FALSE,
computable = FALSE,
at_boundary_ok = TRUE,
all_dists = FALSE,
min_pmix = ssd_min_pmix(nrow(data)),
range_shape1 = c(0.05, 20),
range_shape2 = range_shape1,
control = list(),
silent = FALSE
)Arguments
- data
A data frame.
- left
A string of the column in data with the concentrations.
- ...
Unused.
- right
A string of the column in data with the right concentration values.
- weight
A string of the numeric column in data with positive weights less than or equal to 1,000 or NULL.
- dists
A character vector of the distribution names.
- nrow
A positive whole number of the minimum number of non-missing rows.
- rescale
A flag specifying whether to leave the values unchanged (FALSE) or to rescale concentration values by dividing by the geometric mean of the minimum and maximum positive finite values (TRUE) or a string specifying whether to leave the values unchanged ("no") or to rescale concentration values by dividing by the geometric mean of the minimum and maximum positive finite values ("geomean") or to logistically transform ("odds").
- odds_max
A number specifying the upper left value when
rescale = "odds". By default left values cannot exceed 0.999.- reweight
A flag specifying whether to reweight weights by dividing by the largest weight.
- computable
A flag specifying whether to only return fits with numerically computable standard errors.
- at_boundary_ok
A flag specifying whether a model with one or more parameters at the boundary should be considered to have converged (default = TRUE).
- all_dists
A flag specifying whether all the named distributions must fit successfully.
- min_pmix
A number between 0 and 0.5 specifying the minimum proportion in mixture models.
- range_shape1
A numeric vector of length two of the lower and upper bounds for the shape1 parameter.
- range_shape2
shape2 parameter.
- control
A list of control parameters passed to
stats::optim().- silent
A flag indicating whether fits should fail silently.
Details
By default the 'gamma', 'lgumbel', 'llogis', 'lnorm', 'lnorm_lnorm' and
'weibull' distributions are fitted to the data.
For a complete list of the distributions that are currently implemented in
ssdtools see ssd_dists_all().
If weight specifies a column in the data frame with positive numbers, weighted estimation occurs. However, currently only the resultant parameter estimates are available.
If the right argument is different to the left argument
then the data are considered to be censored.
Examples
fits <- ssd_fit_dists(ssddata::ccme_boron)
fits
#> Distribution 'gamma'
#> scale 25.1268
#> shape 0.950179
#>
#> Distribution 'lgumbel'
#> locationlog 1.92263
#> scalelog 1.23224
#>
#> Distribution 'llogis'
#> locationlog 2.62628
#> scalelog 0.740426
#>
#> Distribution 'lnorm'
#> meanlog 2.56165
#> sdlog 1.24154
#>
#> Distribution 'lnorm_lnorm'
#> meanlog1 0.949483
#> meanlog2 3.20102
#> pmix 0.283968
#> sdlog1 0.554465
#> sdlog2 0.768862
#>
#> Distribution 'weibull'
#> scale 23.514
#> shape 0.9661
#>
#> Parameters estimated from 28 rows of data.
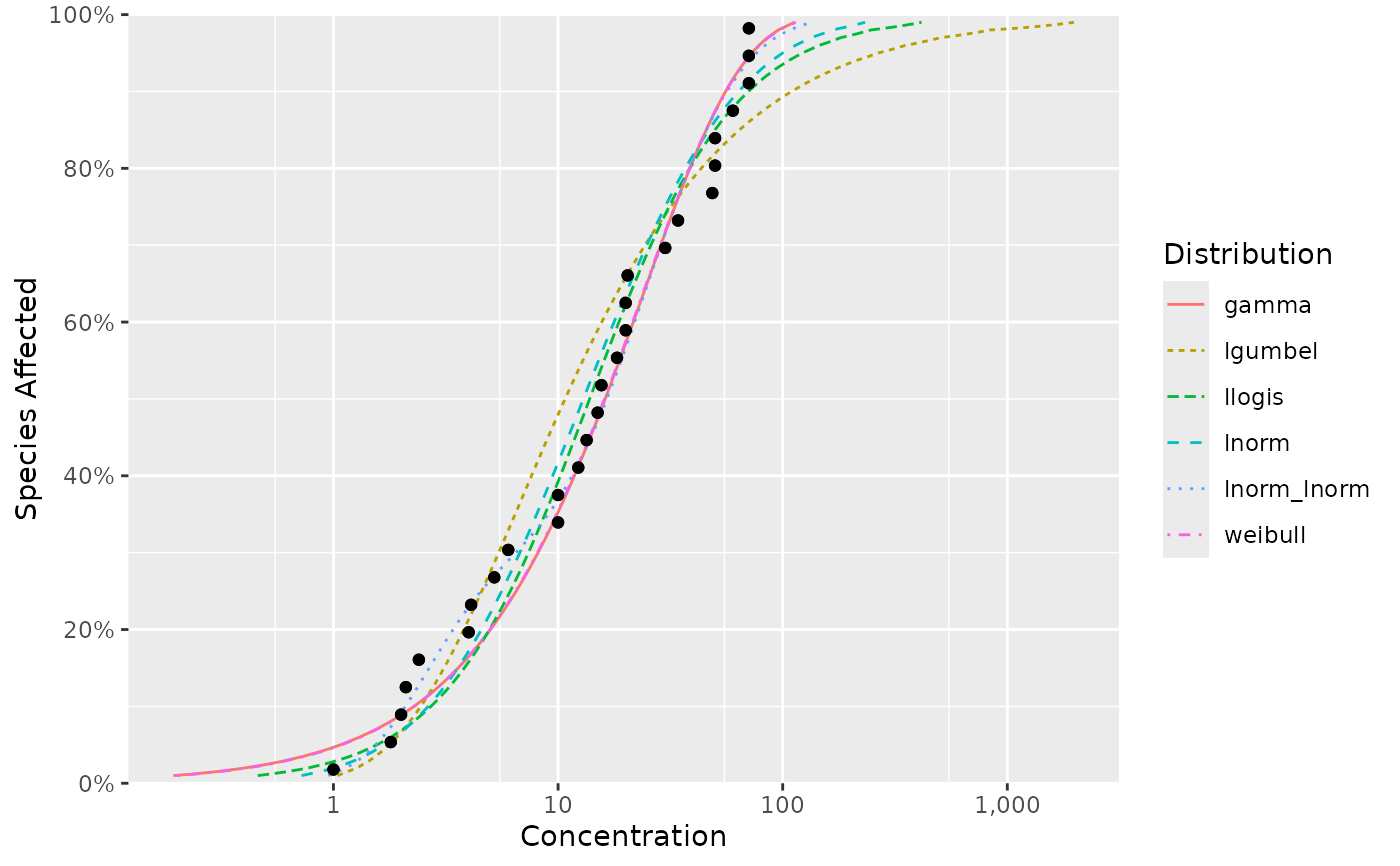
ssd_plot_cdf(fits)
 ssd_hc(fits)
#> # A tibble: 1 × 15
#> dist proportion est se lcl ucl wt level est_method ci_method
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 average 0.05 1.26 NA NA NA 1 0.95 multi weighted_sa…
#> # ℹ 5 more variables: boot_method <chr>, nboot <int>, pboot <dbl>,
#> # dists <list>, samples <list>
ssd_hc(fits)
#> # A tibble: 1 × 15
#> dist proportion est se lcl ucl wt level est_method ci_method
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 average 0.05 1.26 NA NA NA 1 0.95 multi weighted_sa…
#> # ℹ 5 more variables: boot_method <chr>, nboot <int>, pboot <dbl>,
#> # dists <list>, samples <list>
