PEMprepr
Example workflow
PEMprepr.RmdPEMprepr, short for Predictive Ecosystem Mapping –
Preparation is the first of a series of packages supporting the
British Columbia Biogeoecosystem Classification’s
Predictive Ecosystem Mapping (PEM) project. See BC-BEC and PEMr on
_GitHub_for details about the BC BEC classification system and, the PEM
Project.
A portion of the Aleza Lake Research Forest, one of the pilot study sites for this project, is used for this example. The area outlined in red will be used in the steps below
In the example below it is assumed that you are running the following within a new R project.
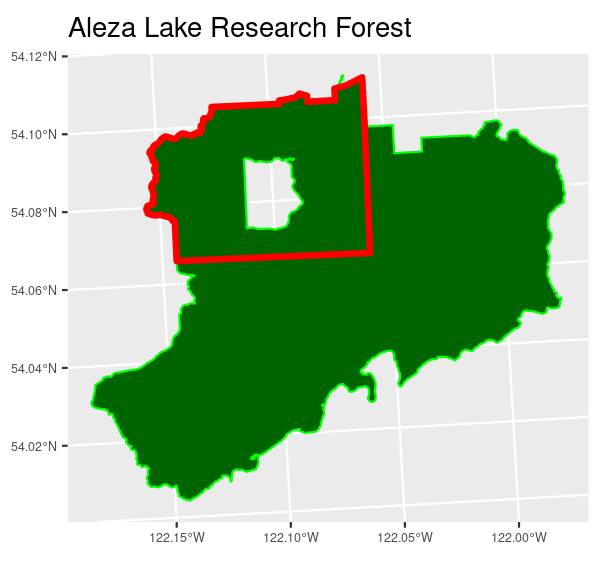
The Aleza Lake Research forest with the area of interest outlined in red
Workflow
The following provides a standard workflow for preparing data for subsequent sampling, modeling, and mapping. The key steps demonstrated below include:
- Data folder set up.
- Alignment of the area of interest
- Generation of covariates
- Creation of a multi-resolution raster stack
- Collection of vector data.
Folder setup
Data for the project can quickly become difficult to manage. To
facilitate ease of data management a standard set of folders for each
area of interest. The following creates the needed folders for the area
of interest: aleza. In addition, fstr stores a
named list of the folders created which we will utilize later (e.g. to
programmatically place results of functions in the correct place).
_Note: if the folders have been generated previously this function will still return the list of folder names.
# creates dirs and returns a named list
fstr <- setup_folders("aleza",
full_names = FALSE) # relative or full path
str(fstr[1:5]) ## first 5 in the list.
#> [1] "The aleza folder already exists - returning folder names."
#> List of 5
#> $ AOI_dir : chr "aleza"
#> $ raw_dir : chr "aleza/00_raw_inputs"
#> $ shape_raw_dir: chr "aleza/00_raw_inputs/vector/raw"
#> $ derived_dir : chr "aleza/10_map_inputs"
#> $ cov_dir : chr "aleza/10_clean_inputs/covariates"Alignment of the area of interest
Load the spatial data
aoi <- sf::st_read(system.file("extdata/aleza_nw.gpkg",
package = "PEMprepr"), quiet = TRUE)
sf::st_bbox(aoi) ## note extent
#> xmin ymin xmax ymax
#> 1250797 1013823 1257215 1019097One concern, with this area-of-interest (aoi)
(red area) is that the bounding box or geographic extent of the area
ends with very random numbers (output above). Using this extent will
cause significant problems later when we attempt to stack numerous
rasters.
Snap the extent
Here the aoi is snapped to an extent divisible by 100.
This will facilitate the generation of rasters data that all fits this
grid extent and thereby can be stacked with relative ease.
aoi <- aoi_snap(aoi, method = "shrink") ## or expand
#> [1] "initial extent is:"
#> xmin ymin xmax ymax
#> 1250797 1013823 1257215 1019097
#> [1] "Extent is:"
#> xmin ymin xmax ymax
#> 1250800 1013900 1257200 1019000Raster templates
The snapped aoi can now be used to create a raster
template. In the example below a 25m2 raster template is
created.
t25 <- create_template(aoi, 25)
t25 ## note that the extent of the raster is the same as the aoi.
#> class : SpatRaster
#> dimensions : 204, 256, 1 (nrow, ncol, nlyr)
#> resolution : 25, 25 (x, y)
#> extent : 1250800, 1257200, 1013900, 1019000 (xmin, xmax, ymin, ymax)
#> coord. ref. : NAD83 / BC Albers (EPSG:3005)
#> source : memory
#> name : lyr.1
#> min value : 0
#> max value : 0Generation of covariates
Here we generate the terrain derived covariates. This is powered by SAGA-GIS. Covariates are generated at multiple resolutions, for example 5, 10, and 25m2.
Steps to create the covariates
- load a digital terrain model (
dtm). Generally, this will be a high resolution model derived from lidar. - align the
dtmto the template. resample thedtmto the desired resolution – here we demonstrate using the 25m2 templated created above. - create the covariates
Import the terrain model
dtm <- terra::rast(system.file("extdata/aleza_nw.tif",
package = "PEMprepr"))
dtm
#> class : SpatRaster
#> dimensions : 1170, 1408, 1 (nrow, ncol, nlyr)
#> resolution : 5.000284, 5.000422 (x, y)
#> extent : 1250480, 1257520, 1013524, 1019375 (xmin, xmax, ymin, ymax)
#> coord. ref. : NAD83 / BC Albers (EPSG:3005)
#> source : aleza_nw.tif
#> name : aleza_nw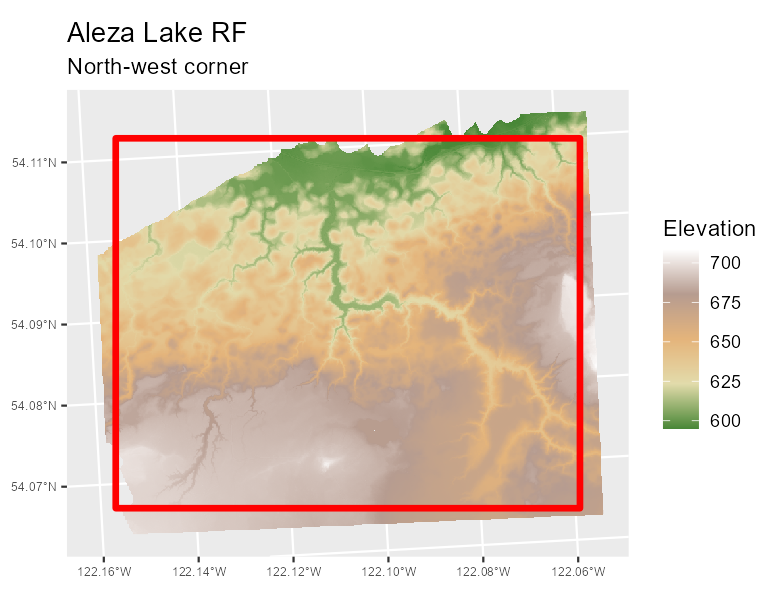
A raw digital terrain model with the area-of-interest noted in red. All processed rasters will align with the aoi.
Align the raster
dtm25 <- align_raster(dtm, t25)
dtm25
#> class : SpatRaster
#> dimensions : 204, 256, 1 (nrow, ncol, nlyr)
#> resolution : 25, 25 (x, y)
#> extent : 1250800, 1257200, 1013900, 1019000 (xmin, xmax, ymin, ymax)
#> coord. ref. : NAD83 / BC Albers (EPSG:3005)
#> source : memory
#> name : aleza_nw
#> min value : 595.0263
#> max value : 704.1022Create the covariates
layer_options <-
c("Filled_sinks", "sinkroute", "dem_preproc",
"slope_aspect_curve", "tCatchment", "tca", "sCatchment",
"twi", "channelsNetwork", "Distance2Water",
"MultiResFlatness", "MultiResFlatness2",
"MultiResFlatness3", "TRI", "convergence", "Openness",
"dah", "TPI", "RidgeValley", "MRN", "FlowAccumulation",
"SlopeLength", "FlowAccumulation2", "FlowAccumulation3",
"FlowPathLength", "FlowPathLength2", "FlowPathLength3",
"LSFactor", "SolarRad", "Convexity", "VertDistance", "TCI_low",
"SWI", "WindExp", "Texture", "Protection", "VRM",
"MBI", "mscale_TPI", "RelPosition", "SlopeCurvatures",
"SteepestSlope", "UpslopeArea")
create_covariates(dtm = dtm25, ## raster created above
SAGApath = "c:/SAGA/", ## Where SAGA GIS is installed
output = fstr$cov_dir, ## from the setup_folders above
layers = "all") ## use all or one of the aboveNote that the pre-defined output directory fstr$cov_dir,
created using setup_folders(), is used to ensure generated
covariates are saved to the correct location. Rasters are also saved
into subfolders of the same resolution.
l <- list.files(path = fstr$cov_dir, pattern = ".sdat",
recursive = TRUE)
l <- l[!grepl("xml",l)]
## created covariates
head(l, 20)
#> [1] "25/aspect.sdat"
#> [2] "25/cnetwork.sdat"
#> [3] "25/convergence.sdat"
#> [4] "25/convexity.sdat"
#> [5] "25/dah.sdat"
#> [6] "25/dem_preproc.sdat"
#> [7] "25/diffinso.sdat"
#> [8] "25/direinso.sdat"
#> [9] "25/down_curv.sdat"
#> [10] "25/dtm.sdat"
#> [11] "25/Filled_sinks.sdat"
#> [12] "25/flow_accum_ft.sdat"
#> [13] "25/flow_accum_p.sdat"
#> [14] "25/flow_accum_td.sdat"
#> [15] "25/flowlength1.sdat"
#> [16] "25/FlowPathLenTD.sdat"
#> [17] "25/gencurve.sdat"
#> [18] "25/hdistnob.sdat"
#> [19] "25/local_curv.sdat"
#> [20] "25/local_downslope_curv.sdat".sdat format – this is read/writable by
terra::rast
Creation of the multi-resolution raster stack
Generation of the covariates above is repeated for each resolution in the multi-resolution model. In order to make all of the rasters stackable, necessary for model and map generation, they must all have the same extent and resolution.
The process above ensures they all have the same extent.
Next the coarse resolution rasters are disaggregated to a fine scale resolution. This means that the data in the resulting raster is the data of the coarse-raster but in a fine scale format.
full.names = TRUE
to list the files.
## we can use the list of files from above
l <- list.files(path = fstr$cov_dir,
pattern = ".sdat",
full.names = TRUE, ## Use TRUE
recursive = TRUE)
l <- l[!grepl("xml",l)]
create_fine_res(inputFileList = l[1:5], ## using the list of files above
output = fstr$cov_dir, ## from setup_folders())
targetRes = 5)
#> [1] "Processing: aleza/10_clean_inputs/covariates/25/aspect.sdat"
#> [1] "Created: aleza/10_clean_inputs/covariates/5/aspect_25.sdat"
#> [1] "Processing: aleza/10_clean_inputs/covariates/25/cnetwork.sdat"
#> [1] "Created: aleza/10_clean_inputs/covariates/5/cnetwork_25.sdat"
#> [1] "Processing: aleza/10_clean_inputs/covariates/25/convergence.sdat"
#> [1] "Created: aleza/10_clean_inputs/covariates/5/convergence_25.sdat"
#> ...This script is a wrapper for terra::disaggregate.
Collection of vector data
A standard set of Vector data is used for subsequent sample planning
and is based on the aoi. This script collects numerous
vector layers from the BC
Data Catalogue making use of the bcdata
package
create_base_vectors(in_aoi = aoi,
out_path = fstr$shape_raw_dir)files are saved in geopackage format.